人类生存依赖农业生产作为粮食来源,但在长期发展过程中,大量病原菌引发的植物病害对粮食生产和粮食安全构成了巨大威胁。尽管目前喷施农药可以控制大多数病原菌造成的病害,但农药的使用对人类健康及环境产生了不利影响。因此,培育具有广谱持久抗性的作物品种成为控制植物病害有效且可持续的方法[1-2]。抗病基因(resistance gene, R)作为培育抗病品种的遗传基础,其通过识别病原菌分泌的无毒基因(avirulence gene, Avr)激活植物效应分子触发的免疫反应(effector-triggered immunity, ETI)。1905年,研究人员首次在小麦中报道了R基因,并由此开启了对R基因在抗病功能方面的遗传研究[3]。20世纪50年代,根据亚麻对亚麻锈菌不同生理小种的特异性抗性表现,Flor[4]提出了“基因对基因(gene for gene)假说”,即携带R基因的寄主植物与携带对应Avr基因的病原微生物互作时,表现为抗病。1994年,科学家在本氏烟中克隆到第一个植物NLR (nucleotide-binding domain and leucine-rich repeat receptor,含核苷酸结合结构域和亮氨酸富集重复区受体蛋白)类抗病基因N,同年拟南芥NLR类抗病基因RPS2也被克隆。烟草N基因通过识别病毒激发子p50从而激活对烟草花叶病毒的抗性,拟南芥RPS2通过识别AvrRpt2激活植物对丁香假单胞菌的抗性[5-6]。迄今为止,至少已有400个抗病基因被鉴定并克隆,其中多个抗病基因已应用于抗病品种的培育并表现出良好效果[7-11]。
根据编码蛋白质的氨基酸基序以及是否含有跨膜结构域,抗病基因可分为不同的家族(表 1),约70%已报道的抗病基因编码NLR。典型的NLR蛋白由N端CC/TIR结构域、中间核苷酸结合结构域(NB)以及C末端的富亮氨酸重复序列(LRR)构成 [12-13]。除CC及TIR外,还有一类NLR基因的N端结构是RPW8型,这类NLR抗病基因包括NRG1和ADR1两类基因家族[14]。在进化过程中,部分NLR蛋白获得了新的集成型结构域(integrated domains, ID),该结构域参与识别病原菌分泌的效应因子进而激活下游免疫反应[15-17]。另外,植物基因组中还存在编码缺失N端或C端部分典型结构域的NLR抗病基因[18]。除NLR类抗病基因外,多种非NLR类抗病基因已被报道(表 1),如番茄抗病基因Pto编码一个典型的丝氨酸/苏氨酸激酶(serine/threonine kinase, STK)[19];番茄抗叶霉病基因Cf-2、Cf-4、Cf-5、Cf-9编码质膜定位的LRR-TM类蛋白,由N端胞外LRR结构域和C端跨膜域(transmembrane, TM)构成[20-23];小麦抗叶枯病基因Stb6编码一个细胞壁类受体激酶(wall-associated receptor kinase, WAK);小麦抗白粉病基因Pm24编码一个串联激酶(tandem kinase-pseudokinase, TKP)[24-30]。
| 表 1 抗病基因(R)结构域分类 |
通常,植物抗病与生长发育均被认为是耗能过程,植物抗病反应的增强会影响自身的生长发育,这种现象也被称为抗病与生长发育的拮抗。在无病原菌侵染的情况下,抗病基因通常仅维持较低的表达水平并且处于活性抑制状态,过量表达或激活均会对生长发育产生不利影响。例如,在拟南芥SNC1功能获得性突变体中,抗病基因表达水平及活性增加导致植株生长矮小、叶片皱缩、结实率下降;持续过量表达辣椒抗病基因Bs3或大麦抗病基因Rph3会导致植物细胞程序性死亡[31-33]。然而,抗病基因的积累不足会导致植物无法激活对病原菌的抗性。例如,马铃薯抗病基因RB的转录水平与其介导的晚疫病抗性之间存在正相关[34]。因此,在与病原菌长期互作过程中,植物进化出复杂且精细的调控网络,在转录水平、转录后水平以及蛋白质水平上精细控制抗病基因的表达与活性,从而平衡抗病与生长发育(图 1)[35-36]。本文将主要围绕NLR类抗病基因表达与活性的调控机制进行探讨,包括转录因子与顺式作用元件、表观遗传修饰、mRNA的选择性剪切和选择性多聚腺苷酸化、sRNA介导的基因沉默,以及蛋白质水平上的泛素化和磷酸化等修饰与调控,并展望这些机制在未来作物抗病遗传育种中的应用前景。
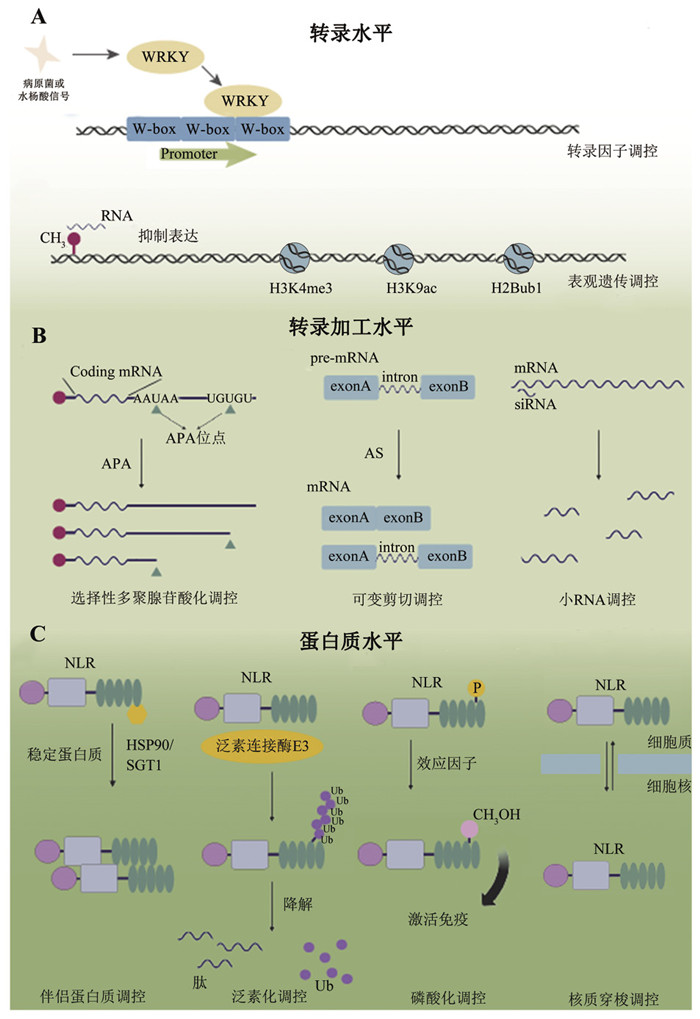
|
图 1 抗病基因调控方式模式图 |
抗病基因的组织特异性及时空特异性表达与其功能紧密相关[37]。转录水平上的调控主要包括转录因子(transcription factors)以及与转录因子结合的顺式作用元件(cis-acting element)。除此之外,包括DNA修饰(DNA甲基化)、组蛋白修饰(组蛋白甲基化、乙酰化、泛素化)以及染色质重塑在内的表观遗传修饰也在抗病基因的转录调控上发挥重要作用。
1.1 转录因子调控转录因子通过与顺式作用元件特异性结合调节基因转录活性,在植物响应病原菌侵染过程中起着至关重要的作用。WRKY家族是植物最大的转录因子家族之一,能够特异性结合下游基因启动子W-box顺式作用元件调控基因转录水平,在植物免疫中发挥重要作用[38]。Mohr等[39]发现W-box元件在植物抗病基因启动子序列中显著富集,其中拟南芥抗病基因RPP8的启动子区域存在三个W-box基序,并且该基因的表达受到病原菌及水杨酸的诱导。当突变三个W-box调控功能域后,抗病基因RPP8的表达水平无法被病原菌诱导上调,导致植物抗病性下降,表明抗病基因RPP8启动子区W-box基序对于其表达水平及抗性激活具有重要作用,同时W-box在抗病基因中的保守存在暗示了WRKY转录因子对于抗病基因表达以及免疫调控的潜在意义[39]。锌指类转录因子在抗病基因转录调控中也发挥重要作用。水稻锌指蛋白OsVOZ1和OsVOZ2可结合并促进抗病基因Piz-t的表达,以提高Piz-t介导的稻瘟病抗性[40]。拟南芥抗病基因SNC1的精细表达对生长和免疫的平衡具有重要意义,通过筛选snc1突变体的抑制子获得了多个mos (modifier of snc1) 突变体,发现MOS1通过与转录因子TCP15结合增强其与SNC1启动子的结合进而促进SNC1的表达。除转录因子之外,多个位于启动子区和下游非编码区的顺式作用元件也共同调控SNC1的表达[41-42]。
由于抗病基因的转录对激活抗病反应至关重要,因而在与病原菌的“军备竞赛”中,植物抗病基因进化出一种“诱饵型”顺式作用元件,用于快速响应病原菌侵染。TALE (TAL effector)是一类最初从黄单胞菌(Xanthomonas spp.)中鉴定到的转录因子类效应分子,通过结合并促进植物感病基因的表达,以提高病原菌的致病力。相应地,植物抗病基因启动子区域进化出一类特定基序,作为“诱饵”被TALE结合并促进抗病基因表达。例如,辣椒抗病基因Bs3启动子区域进化出黄单胞菌TAL效应分子AvrBs3的结合基序,黄单胞菌分泌的AvrBs3进入植物细胞,结合并激活Bs3的表达,从而引发植物抗病反应[32]。目前,已报道多个抗病基因的表达和激活受到病原菌分泌的TAL效应分子的转录调控,如大麦抗病基因Rph3[31],水稻白叶枯抗病基因Xa10、Xa23、Xa27[43-45],以及辣椒抗病基因Bs4C等[46]。
1.2 表观遗传调控DNA甲基化是表观遗传水平上的修饰之一,这一修饰存在于所有高等生物中,并与基因表达调控密切相关。研究发现植物抗病基因广泛存在DNA甲基化修饰,并且该修饰参与调控植物抗病反应[47]。全基因组甲基化水平分析发现,菜豆NLR抗病基因中约一半存在甲基化修饰,远高于其他基因家族。其中,约有90个被24-nt的siRNA靶向,且这些NLR基因的表达水平很低甚至不表达,表明其受RdDM (RNA介导的DNA甲基化)介导的基因沉默调控[48]。除全基因组水平外,一些抗病基因的表达调控也与DNA甲基化修饰高度相关。水稻抗病基因PigmR能够为稻瘟病不同生理小种提供广谱抗性,但过量表达或持续激活严重影响水稻生长与产量,同一位点的PigmS能够与PigmR互作形成异源二聚体并抑制PigmR介导的抗病反应。研究发现PigmS的组织特异性表达受RdDM调控,叶片中PigmS启动子区域存在较高的甲基化水平并且表达量较低,而在花粉中该基因的甲基化水平较低,因此表达量较高,通过调控不同组织中Pigm的表达水平可实现水稻产量和抗病之间的平衡[10]。水稻抗病基因Xa21G启动子区域存在5mC甲基化修饰,导致其无法表达,而在人工去甲基化处理的突变体中,消除Xa21G启动子区域的甲基化能恢复其自身表达量,表现出对黄单胞菌水稻致病变种的抗性[49]。5mC甲基化修饰对抗病基因的表达也具有调控功能,如水稻抗稻瘟病基因Pib启动子区域5mC甲基化修饰与Pib的表达水平呈正相关[50]。因此,不同类型的甲基化修饰对抗病基因的表达及功能具有重要的调控作用,高水平的甲基化往往能够降低抗病基因的转录水平,减少细胞的能量消耗,从而维持正常的生长;相反,当病原菌攻击时抗病基因也能通过甲基化调控增加自身表达量,快速激活植物对病原菌的抗性[51]。
组蛋白作为核小体的基本组成成分能够发生多种化学修饰,如甲基化、乙酰化、泛素化等。这些修饰通过调控染色质的状态进而调控基因的表达及稳定性,在植物的生长发育及外界信号响应过程中具有重要作用[52]。例如,H3K4甲基转移酶ATXR7通过调控抗病基因SNC1以及RPP4上的H3K4me3水平增强它们的表达,进而激活拟南芥对霜霉病的抗性[53]。组蛋白甲基转移酶SDG8参与调控拟南芥抗病基因LAZ5的H3K36me3修饰水平以持续激活LAZ5转录活性,进而影响植物免疫[54-56]。高粱抗病基因ARG1 5'末端非翻译区(5'UTR)的H3K36me2及H3K36me3在病原菌侵染过程中显著富集,导致ARG表达水平上调从而提高高粱的抗性水平;而在高粱感病品种中,ARG1基因上H3K36me2/me3水平较低,导致基因表达水平较低,无法对病原菌产生完全抗性[57]。组蛋白乙酰化对NLR基因的表达也具有调控作用,例如,组蛋白去乙酰化酶(HDACs) HDA9与其互作蛋白HOS15能够调控多个拟南芥抗病基因的H3K9乙酰化修饰水平,进而调控抗病基因表达以达到抗病与生长发育平衡[58]。除此之外,E3泛素连接酶HUB1和HUB2介导SNC1位点上的H2B泛素化,在病原菌侵染时促进SNC1的表达,并与组蛋白甲基化和乙酰化共同调控SNC1的表达[59-60]。
染色质重塑发生在基因复制和重组过程中,是指染色质包装状态、核小体中组蛋白以及对应DNA分子发生的改变。染色质重塑复合物由染色质重塑因子组成,分为四大类:SWI/SNF、CHD、ISWI和INO80[61]。研究发现染色质重塑复合物对于抗病基因的表达具有重要调节作用。BAF60是SWI/SNF染色质重塑复合物中的一个亚基,可通过ATP水解产生的能量在DNA上定位核小体[62]。Huang等[63]发现BAF60在拟南芥中的同源蛋白SWP73A能够结合抗病基因RPS2启动子区域调控H3K9me2水平以抑制RPS2的表达;当病原菌侵染时,miR3440和siRNA-SWP73A靶向SWP73A导致该基因沉默,因此激活了RPS2的表达以及拟南芥的抗病反应。Zou等[64]发现染色质重塑蛋白CHR5作为CHD染色质重塑复合体的亚基,可调控核小体占有率,参与调控SNC1的表达水平。此外,拟南芥SWI/SNF染色质重塑复合体SYD也在染色质水平上抑制SNC1的转录[65]。
2 RNA水平调控基因的表达调控不仅发生在转录水平,在转录后水平上也存在多种调控方式。转录后调控(post-transcriptional regulation)是指发生在RNA水平上的调控作用,具体包括pre-mRNA的多聚腺苷酸化和剪切加工、mRNA的稳定性调控、RNA干扰介导的转录后基因沉默等[66]。
2.1 选择性多聚腺苷酸化调控转录后mRNA在polyA加尾时可能会选取不同的位置,使得一个基因可产生不同3'UTR的转录本,也被称为选择性多聚腺苷酸化(alternative polyadenylation, APA)。APA普遍存在于真核生物中,参与调控基因转录本的多样性和稳定性。研究发现,拟南芥开花相关基因FLC、FPA及FCA的表达水平都受到APA的调控[67]。抗病基因表达模式分析发现在拟南芥72个抗病基因上都鉴定到APA,表明该调控在抗病基因上普遍存在,但具体功能还不清楚[37]。2017年,Tsuchiya等[68]发现RPP7的5'UTR区域插入了一个转座子,该转座子插入位置携带H3K9me2修饰,这种甲基化修饰水平的变化导致RPP7的多聚腺苷酸化位点发生改变;遗传分析发现,EDM2蛋白参与了RPP7基因的选择性多聚腺苷酸化,直接调控RPP7具有抗性功能的转录本的表达水平,对其介导的抗病反应至关重要。
2.2 前体RNA加工调控RNA剪切是指前体RNA (pre-mRNA)中内含子去除、外显子连接的过程,这一过程由剪切复合体完成。剪切复合体是一种进化上保守的核糖核蛋白复合物,通过识别内含子结构并对其剪切产生成熟的mRNA,这一过程可能会发生可变剪切,导致同一基因产生多种转录本。这种调控方式在植物中普遍存在,已知拟南芥60%的非单个外显子基因均存在可变剪切调控[69],而其他双子叶及单子叶植物中也有30%~50%的多外显子基因存在可变剪切调控[70-72]。
可变剪切对于蛋白质多样性和丰富性具有重要意义[73],大量研究发现植物抗病基因受到可变剪切调控。Mandadi等[74]利用RNA-seq数据对二穗短柄草(Brachypodium distachyon)受病毒侵染过程中的可变剪切事件进行分析,发现100多个免疫相关基因在病毒侵染过程中发生可变剪切,其中包含9个抗病基因。研究人员对拟南芥剪切因子SR45的功能进行分析,发现包括多个抗病基因在内的542个基因发生的可变剪切受到SR45的调控,因此sr45-1突变体植物表现出增强的抗病表型可能与这些抗病基因的表达水平相关[75]。这些研究表明植物抗病基因可变剪切广泛存在,并且对抗病基因功能的调控发挥重要作用。
烟草抗病毒基因N是植物中第一个被报道的存在可变剪切的抗病基因,可变剪切产生的编码全长蛋白的转录本NS和编码截短蛋白的转录本NL共同激活烟草对烟草花叶病毒的抗性[76]。拟南芥RPS4可变剪切产生的其中一条转录本编码一种截短蛋白,该蛋白能够自激活引发植物细胞坏死[77]。RPS4通过可变剪切调控多个转录本的表达以抑制自激活,维持植物正常生长发育。早期鉴定到的抗病基因可变剪切事件大多发生在TNL (TIR-NB-LRR)类抗病基因中,这主要源于TNL类基因含有较多内含子结构,增加了可变剪切发生的可能性。研究发现CNL (CC-NB-LRR)类抗病基因也存在可变剪切现象,并且不同转录本在植物免疫中发挥不同的功能。大麦CNL基因Mla13的上游开放阅读框(uORF)区域含有两个内含子并通过可变剪切产生5种转录本,在病原菌侵染过程中,Mla13内含子剪切转录本转录上调,激活大麦抗性[78];水稻CNL基因Pi-ta能够发生可变剪切,产生11种转录本,其中5个转录本正常表达并编码不同的蛋白,病原菌侵染时抗病品种中编码TPX结构域蛋白的转录本表达水平升高[79];小麦NLR基因Lr10发生可变剪切产生两种转录本,并且同时含有这两种转录本的硬粒小麦对叶锈病表现抗性[80];水稻抗病基因RGA5通过内含子保留形式产生两种转录本,而发生可变剪切的区域包含抗病基因识别其无毒基因Avr1-CO39的关键区段,导致内含子保留转录本编码的抗病蛋白无法识别Avr1-CO39 [81]。
目前,NLR基因的可变剪切调控机制还未被完全解析。Xu等[82]首次发现植物一类富含丝氨酸/精氨酸蛋白家族(serine/arginine-rich protein, SR)中的MOS12功能缺失会影响拟南芥抗病基因SNC1及RPS4的剪切模式。马铃薯广谱抗病基因RB的转录本表达量在侵染过程中受其无毒基因AVRblb1调控,同时马铃薯中保守的剪切因子CWC15参与可变剪切调控过程,进而影响RB介导的晚疫病抗性[83]。无义介导的mRNA降解(nonsense-mediated mRNA decay, NMD)作为一种对可变剪切产物加工的机制,通过识别并降解含有提前终止密码子的mRNA,避免产生截短的蛋白产物来对细胞产生毒性[84]。NMD对抗病基因表达也具有调控作用,研究发现植物NMD复合物的靶标中近三分之一是抗病基因[85]。例如,NMD突变体中RPS6的内含子剪切转录本会过量积累,导致植物生长缺陷[86-87]。
2.3 小RNA调控植物小RNA (small RNA, sRNA)参与调控生长发育及胁迫响应等多种生命活动,是基因表达的重要调控因子。sRNA主要分为两类:微小RNA (microRNA, miRNA)以及小干扰RNA (small interfering RNA, siRNA)。sRNA主要通过与靶标mRNA序列反向互补抑制基因表达,或通过对mRNA进行切割降解mRNA或抑制翻译[88]。植物抗病基因的表达水平受到不同miRNA及siRNA的调控作用。Yi和Richards [89]首次发现拟南芥中的sRNA参与调控抗病基因的表达,其研究表明包括21-nt siRNA在内的多个siRNA共同作用抑制RPP5基因簇上包括SNC1及RPP4在内的多个抗病基因的表达。在其他植物中发现一些miRNA基因家族参与调控抗病基因的表达。例如,蒺藜苜蓿(Medicago truncatula)中3个miRNA家族靶向多达74个抗病基因,并进一步生成具有相位排列结构特征的植物内源siRNA (phase secondary small interfering RNA, phasiRNA),这类小RNA通过与AGO蛋白结合对靶标mRNA进行切割降解,最终抑制抗病基因的表达[90]。有研究报道phasiRNA在豆类及茄科植物中也能够靶向抗病基因,并导致其转录后沉默[91]。因此,通过编辑植物内源sRNA调控抗病基因的精细表达,可为平衡抗病与生长发育提供新策略[92]。
3 蛋白质水平调控抗病基因在蛋白质水平的调控是抗病基因功能调节的重要组成部分,其主要分为蛋白质的加工和修饰。加工是指抗病基因翻译后的产物伴随其他蛋白一起组成功能蛋白,这些蛋白质往往被称作伴侣蛋白,而修饰则指通过在氨基酸残基上发生的磷酸化、乙酰化等化学修饰调控抗病蛋白的功能。
3.1 分子伴侣和辅助伴侣分子调控与动物中的抗病蛋白相似,植物抗病蛋白也需要其他蛋白来促进其折叠。一类热休克蛋白能够调控蛋白质成熟以及降解错误折叠的多肽,因而影响多个信号转导蛋白的活性,这种蛋白也被称为伴侣蛋白[93]。植物伴侣蛋白HSP90参与调控多个抗病蛋白的稳定性,HSP90的突变或沉默降低了抗病蛋白Rx、RPM1以及RPS5的表达水平[94-96]。同时,遗传分析表明HSP90介导的蛋白质稳定性直接参与调控抗病蛋白介导的免疫反应[97]。此外,研究发现植物中保守的伴侣蛋白RAR1和SGT1能够与HSP90发生互作,形成复合体参与调节多个抗病蛋白的稳定性。同时突变HSP90、SGT1以及RAR1时,包括N、RPS2在内的多个抗病蛋白表达下降,导致免疫响应降低[98]。单独突变RAR1时,拟南芥RPM1和RPS5、大麦Mla1和Mla6以及马铃薯Rx等多个抗病蛋白的表达均受到影响[96, 99-100]。同样,SGT1的缺失也造成多个抗病蛋白的积累下降,如Rx以及N [101-103]。
3.2 泛素化调控机制蛋白质泛素化是一种常见的蛋白质翻译后修饰(post-translational modification),主要通过泛素蛋白酶体系统(ubiquitin proteasome system, UPS)调控蛋白质的降解进而影响其功能。植物抗病蛋白受到复杂的泛素化调控,以确保激活后的抗病蛋白在发挥抗病功能后被正确降解,以避免过高的抗病反应抑制生长发育。目前,已有的研究表明E3泛素连接酶是调控植物抗病蛋白泛素化的主要蛋白,E3连接复合酶体SCF (SKP1/Cullin1/F-box)中的F-box蛋白CPR1调控拟南芥抗病蛋白SNC1、RPPS2和SUMM2的降解,其缺失会导致这些蛋白质持续积累[33, 104-105]。此外,拟南芥中抗病蛋白SIKIC2的表达受到两个E3连接酶MUSE1和MUSE2的负调控[106]。番茄E3泛素连接酶SBP1与番茄抗病蛋白Sw-5b互作并介导其降解[107]。烟草E3泛素连接酶UBR7与抗病N蛋白互作,UBR7表达下调导致N蛋白表达水平升高进而增强烟草对TMV的抗性水平[108]。尽管目前对抗病蛋白的泛素化修饰的研究取得了很大进展,但鉴于抗病蛋白的分子量较大,结构复杂,更多翻译后修饰在抗病蛋白上的作用机制还没有明确报道。因此,挖掘抗病蛋白发生的修饰并解析其功能机制对理解抗病蛋白的作用机制有重要意义,可为更好地应用抗病蛋白提供理论基础。
3.3 磷酸化调控机制蛋白质磷酸化修饰是调控细胞信号转导的主要机制,其通过蛋白激酶的催化作用将磷酸基团转移到蛋白质的氨基酸残基上,如丝氨酸、苏氨酸以及酪氨酸[109]。蛋白质磷酸化在PTI (pattern-triggered immunity)过程中发挥重要作用。当模式识别受体(pattern recognition receptor, PRR)识别到病原体相关分子模式(pathogen-associated molecular pattern, PAMP)信号分子后,与共受体激酶BAK1互作并相互磷酸化,以迅速激活下游免疫反应[110]。研究发现磷酸化对抗病蛋白的激活具有重要意义,例如,拟南芥成对抗病蛋白RRS1和RPS4可以识别青枯雷尔氏菌分泌的效应因子PopP2以及丁香假单胞效应分子AvrRPS4,激活植物对不同病原菌的抗性[111-113]。RRS1在拟南芥中存在等位变异,其中RRS1-S仅识别AvrRPS4,而RRS1-R可同时识别AvrRPS4和PopP2。序列分析发现,相较于RRS1-S,RRS1-R末端第1 214位苏氨酸可发生磷酸化,且该磷酸化可抑制其自身活性。PopP2编码一个乙酰转移酶,可将RRS1-R末端磷酸化位点乙酰化,从而解除对RRS1-R的抑制,激活下游免疫响应[114]。该研究解析了不同蛋白质修饰对同一抗病蛋白活性的调控机制,同时也展示了一个植物抗病蛋白在进化上利用自身蛋白修饰位点诱捕病菌效应蛋白的新策略。
3.4 核质穿梭调控抗病蛋白的亚细胞定位与其功能息息相关。目前,已报道的抗病蛋白主要在细胞膜、细胞质及细胞核中发挥功能。研究发现,部分抗病蛋白具有质核定位的变化,并且该变化对于抗病蛋白的激活具有重要作用[115]。例如,拟南芥SNC1蛋白在细胞核中积累会造成免疫自激活,而将核输出信号与SNC1融合后可抑制SNC1的自激活,表明核定位对于SNC1介导的免疫激活至关重要[116-118]。进一步研究发现,SNC1的入核受到输入蛋白MOS6的调控,而SNC1的出核受转运蛋白KA120调控[119-120]。部分抗病蛋白在不同亚细胞结构中均发挥功能,但其作用机制存在差异。番茄抗病蛋白Sw-5b分布于细胞质和细胞核中,研究发现定位在胞质中有利于Sw-5b诱导强烈的细胞死亡但无法阻止病毒在细胞间和长距离的移动;而定位在细胞核中的Sw-5b虽触发的免疫较弱,但能有效阻止病毒在细胞间和全系统中的扩散。Sw-5b定位的变化受到其N端SD结构域与转运蛋白互作的调控,表明植物抗病蛋白能够通过核质穿梭调控植物抗性水平[121]。此外,植物抗病蛋白激活后形成抗病小体发挥功能,下游helper NLR蛋白NRG1和ADR1可在细胞膜上形成钙离子通道,激活下游免疫响应;同时,也有研究发现NRG1和ADR1在核里与信号蛋白EDS1形成复合体,参与抗病信号的传递[122-125]。
虽然抗病蛋白亚细胞定位与功能的联系已得到广泛验证,但是其分子机制尚不清晰。除核孔转运蛋白外,核孔复合体蛋白、蛋白质修饰以及蛋白质序列的变化均可能参与抗病蛋白亚细胞定位的变化[126-128]。未来对抗病蛋白调控机制的深入研究有望解析其亚细胞定位变化的调控机理,并为抗病品种的培育提供新思路。
4 抗病基因应用展望随着生物与数据技术的快速发展,植物抗病基因的鉴定和克隆已取得了重要进展,但培育具有农业高价值的作物抗病新品种仍面临着挑战。一方面,需要充分利用抗病基因的持久性及广谱性;另一方面,要在提高抗性的同时,平衡抗病与产量的关系。前期植物抗病小体的发现、抗性形成与调控机制的解析为抗病基因的应用提供了新型理论支撑。随着科技的发展,人工智能在植物抗病育种的各个方面表现出更大潜力,包括数据收集、解锁基因库内的遗传多样性,以及弥合基因型及表型差距以促进作物育种,使开发更适应未来环境的作物品种具有可能性。并且,随着CRISPR/Cas等基因编辑技术的发展以及AlphaFold等生物大分子结构大模型的建立,作物抗病遗传育种已从最初仅依赖于从特异种质资源中挖掘新基因的单一策略,发展形成了包括定向设计和改造原有基因在内的多元化路径。例如,改进后的CRISPR/Cas9系统作为新的基因编辑工具,能够实现更精确和高效的基因组修饰,包括敲除、敲入、基因功能修正或是定向替换DNA片段。这种精准的基因组设计有望成为常规方法,创造出更抗病且高产的品种,并且提高作物的营养价值和增强其适应环境条件的能力。人工智能的发展推动作物育种进入多组学时代,利用整合遗传学、基因组学、转录组学、代谢组学等不同层次的组学数据,可以更深入揭示植物抗病性状和基因组的关系,发现潜在的基因性状关联,预测植物性状和质量并优化育种策略,为培育新的抗病品种提供科学依据。抗病蛋白的设计和改造主要针对NLR,通过随机突变、融合新的ID结构域以及基于结构特征对关键位点进行定点突变等以提高NLR介导的抗病反应,扩大识别谱或创造对效应蛋白的新型识别[129-132]。近年来,通过改造抗病基因的表达模式来创制具有广谱高抗性的作物新种质已成为一个重要的研究方向。例如,利用组成型强启动子结合上游uORF序列共同调控抗病基因的表达,可实现抗病基因快速响应病原菌的侵染,从而在不影响产量的同时,大幅提高作物抗病性[133-134]。利用引导基因编辑技术,将TALE效应因子响应元件定向敲入到水稻抗病基因Xa23的启动子区域中,以此增强Xa23对白叶枯病菌侵染的响应表达水平,从而提高水稻对白叶枯病的抗性。
最后,在利用抗病基因培育抗病品种的过程中,有哪些基因影响抗病和产量平衡?是否能够通过其他元件微调抗病基因表达来平衡植物的抗病和生长?不同遗传背景和环境中的材料抗性如何评价,是否存在适合度代价?这些问题的解决将增加我们对抗病基因的理解,也能为植物的抗病育种提供新的策略以及有效快速地选择高抗且不影响产量的品种,通过精准调控抗病基因来实现广谱抗病基因的长久有效使用将是植物育种的重要思路。未来,利用CRISPR/Cas介导的启动子区域删除替换技术以及蛋白质和RNA修饰关键位点的定点改良等方法,也可利用单细胞组学和空间组学技术追踪抗病基因在不同组织、细胞中的时空表达和调控模式,为作物抗性改良提供更加精准的导向。
| [1] |
Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science, 2013, 341: 746-51. DOI:10.1126/science.1236011 |
| [2] |
Damalas CA, Eleftherohorinos IG. Pesticide exposure, safety issues, and risk assessment indicators. Int J Environ Res Public Health, 2011, 8: 1402-19. DOI:10.3390/ijerph8051402 |
| [3] |
Biffen RH. Mendel's laws of inheritance and wheat breeding. J Agric Sci, 1905, 1: 4-48. DOI:10.1017/S0021859600000137 |
| [4] |
Flor HH. Host-parasite interactions in flax rust-its genetics and other implications. Phytopathology, 1955, 45: 680-5. |
| [5] |
Whitham S, Dinesh-Kumar SP, Choi D, et al. The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin-1 receptor. Cell, 1994, 78: 1101-15. DOI:10.1016/0092-8674(94)90283-6 |
| [6] |
Bent AF, Kunkel BN, Dahlbeck D, et al. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science, 1994, 265: 1856-60. DOI:10.1126/science.8091210 |
| [7] |
Jiorgos K, van der Hoorn RAL. Defended to the Nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell, 2018, 30: 285-99. DOI:10.1105/tpc.17.00579 |
| [8] |
Li W, Deng Y, Ning Y, et al. Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu Rev Plant Biol, 2020, 71: 575-603. DOI:10.1146/annurev-arplant-010720-022215 |
| [9] |
冯晶, 王凤涛, 蔺瑞明, 等. 小麦条锈病抗病遗传及菌源基地基因布局研究进展. 植物保护学报, 2022, 49: 263-75. |
| [10] |
Deng Y, Zhai K, Xie Z, et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science, 2019, 355: 962-5. |
| [11] |
陈子强, 田大刚, 梁廷敏, 等. 229份水稻品种及重要育种材料抗稻瘟病Pik位点基因型鉴定. 福建农业学报, 2016, 31: 553-9. |
| [12] |
Qi D, DeYoung BJ, Innes RW. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol, 2012, 158: 1819-32. DOI:10.1104/pp.112.194035 |
| [13] |
Martin R, Qi T, Zhang H, et al. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science, 2020, 370: eabd9993. DOI:10.1126/science.abd9993 |
| [14] |
Wang H, Song S, Gao S, et al. The NLR immune receptor ADR1 and lipase-like proteins EDS1 and PAD4 mediate stomatal immunity in Nicotiana benthamiana and Arabidopsis. Plant Cell, 2024, 36: 427-46. DOI:10.1093/plcell/koad270 |
| [15] |
Cesari S, Bernoux M, Moncuquet P, et al. A novel conserved mechanism for plant NLR protein pairs: the "integrated decoy" hypothesis. Front Plant Sci, 2014, 5: 606. |
| [16] |
Marchal C, Zhang J, Zhang P, et al. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants, 2018, 4: 662-8. DOI:10.1038/s41477-018-0236-4 |
| [17] |
Bialas A, Langner T, Harant A, et al. Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain. Elife, 2021, 10: e66961. DOI:10.7554/eLife.66961 |
| [18] |
Nishimura MT, Anderson RG, Cherkis KA, et al. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci U S A, 2017, 114: E2053-62. |
| [19] |
Martin GB, Brommonschenkel SH, Chunwongse J, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science, 1993, 262: 1432-6. DOI:10.1126/science.7902614 |
| [20] |
Dixon MS, Jones DA, Keddie JS, et al. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell, 1996, 84: 451-9. DOI:10.1016/S0092-8674(00)81290-8 |
| [21] |
Thomas CM, Jones DA, Parniske M, et al. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell, 1997, 9: 2209-24. |
| [22] |
Dixon MS, Hatzixanthis K, Jones DA, et al. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell, 1998, 10: 1915-25. DOI:10.1105/tpc.10.11.1915 |
| [23] |
Jones DA, Thomas CM, Hammond-Kosack KE, et al. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science, 1994, 266: 789-93. DOI:10.1126/science.7973631 |
| [24] |
Saintenac C, Lee WS, Cambon F, et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat Genet, 2018, 50: 368-74. DOI:10.1038/s41588-018-0051-x |
| [25] |
Kema GHJ, Mirzadi Gohari A, Aouini L, et al. Stress and sexual reproduction affect the dynamics of the wheat pathogen effector AvrStb6 and strobilurin resistance. Nat Genet, 2018, 50: 375-80. DOI:10.1038/s41588-018-0052-9 |
| [26] |
Zhong Z, Marcel TC, Hartmann FE, et al. A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol, 2017, 214: 619-31. DOI:10.1111/nph.14434 |
| [27] |
Klymiuk V, Yaniv E, Huang L, et al. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun, 2018, 9: 3735. DOI:10.1038/s41467-018-06138-9 |
| [28] |
Brueggeman R, Rostoks N, Kudrna D, et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci U S A, 2002, 99: 9328-33. DOI:10.1073/pnas.142284999 |
| [29] |
Chen S, Rouse MN, Zhang W, et al. Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol, 2020, 225: 948-59. DOI:10.1111/nph.16169 |
| [30] |
Lu P, Guo L, Wang Z, et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun, 2020, 11: 680. DOI:10.1038/s41467-020-14294-0 |
| [31] |
Dinh HX, Singh D, Gomez de la Cruz D, et al. The barley leaf rust resistance gene Rph3 encodes a predicted membrane protein and is induced upon infection by avirulent pathotypes of Puccinia hordei. Nat Commun, 2022, 13: 2386. DOI:10.1038/s41467-022-29840-1 |
| [32] |
Römer P, Hahn S, Jordan T, et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science, 2007, 318: 645-8. DOI:10.1126/science.1144958 |
| [33] |
Cheng YT, Li Y, Huang S, et al. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A, 2011, 108: 14694-9. DOI:10.1073/pnas.1105685108 |
| [34] |
James M, Bradeen MI, Dimitre S., et al. Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. MPMI, 2009, 22: 437-46. DOI:10.1094/MPMI-22-4-0437 |
| [35] |
He Z, Webster S, He SY. Growth-defense trade-offs in plants. Curr Biol, 2022, 32: R634-9. DOI:10.1016/j.cub.2022.04.070 |
| [36] |
Huot B, Yao J, Montgomery BL, et al. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant, 2014, 7: 1267-87. DOI:10.1093/mp/ssu049 |
| [37] |
Tan X, Meyers BC, Kozik A, et al. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol, 2007, 7: 56. DOI:10.1186/1471-2229-7-56 |
| [38] |
Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol, 2009, 150: 1648-55. DOI:10.1104/pp.109.138990 |
| [39] |
Mohr TJ, Mammarella ND, Hoff T, et al. The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol Plant Microbe Interact, 2010, 23: 1303-15. DOI:10.1094/MPMI-01-10-0022 |
| [40] |
Wang J, Wang R, Fang H, et al. Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice. Mol Plant, 2021, 14: 253-66. DOI:10.1016/j.molp.2020.11.005 |
| [41] |
Zhang N, Wang Z, Bao Z, et al. MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. Plant J, 2018, 93: 66-78. DOI:10.1111/tpj.13757 |
| [42] |
Yu H, Yang L, Li Z, et al. In situ deletions reveal regulatory components for expression of an intracellular immune receptor gene and its co-expressed genes in Arabidopsis. Plant Cell Environ, 2022, 45: 1862-75. DOI:10.1111/pce.14293 |
| [43] |
Tian D, Wang J, Zeng X, et al. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell, 2014, 26: 497-515. DOI:10.1105/tpc.113.119255 |
| [44] |
Wang C, Zhang X, Fan Y, et al. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant, 2015, 8: 290-302. DOI:10.1016/j.molp.2014.10.010 |
| [45] |
Gu KY, Yang B, Tian DS, et al. R gene expression induced by a type-Ⅲ effector triggers disease resistance in rice. Nature, 2005, 435: 1122-5. DOI:10.1038/nature03630 |
| [46] |
Strauß T, van Poecke RMP, Strauß A, et al. RNA-seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc Natl Acad Sci U S A, 2012, 109: 19480-5. DOI:10.1073/pnas.1212415109 |
| [47] |
Tirnaz S, Batley J. DNA methylation: toward crop disease resistance improvement. Trends Plant Sci, 2019, 24: 1137-50. DOI:10.1016/j.tplants.2019.08.007 |
| [48] |
Richard MMS, Gratias A, Thareau V, et al. Genomic and epigenomic immunity in common bean: the unusual features of NB-LRR gene family. DNA Res, 2018, 25: 161-72. DOI:10.1093/dnares/dsx046 |
| [49] |
Akimoto K, Katakami H, Kim HJ, et al. Epigenetic inheritance in rice plants. Ann Bot, 2007, 100: 205-17. DOI:10.1093/aob/mcm110 |
| [50] |
Li Y, Xia Q, Kou H, et al. Induced Pib expression and resistance to Magnaporthe grisea are compromised by cytosine demethylation at critical promoter regions in rice. J Integr Plant Biol, 2011, 53: 814-23. DOI:10.1111/j.1744-7909.2011.01070.x |
| [51] |
Yaish MW. DNA methylation-associated epigenetic changes in stress tolerance of plants. Mol Stress Physiol Plants, 2013, 427-40. |
| [52] |
Zhang H, Lang Z, Zhu JK. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol, 2018, 19: 489-506. DOI:10.1038/s41580-018-0016-z |
| [53] |
Xia S, Cheng YT, Huang S, et al. Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol, 2013, 162: 1694-705. DOI:10.1104/pp.113.214551 |
| [54] |
Dong G, Ma DP, Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem Biophys Res Commun, 2008, 373: 659-64. DOI:10.1016/j.bbrc.2008.06.096 |
| [55] |
Li Y, Mukherjee I, Thum KE, et al. The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol, 2015, 16: 79. DOI:10.1186/s13059-015-0640-2 |
| [56] |
Palma K, Thorgrimsen S, Malinovsky FG, et al. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog, 2010, 6: e1001137. DOI:10.1371/journal.ppat.1001137 |
| [57] |
Lee S, Fu F, Liao CJ, et al. Broad-spectrum fungal resistance in sorghum is conferred through the complex regulation of an immune receptor gene embedded in a natural antisense transcript. Plant Cell, 2022, 34: 1641-65. DOI:10.1093/plcell/koab305 |
| [58] |
Yang L, Chen X, Wang Z, et al. HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol, 2020, 226: 507-22. DOI:10.1111/nph.16380 |
| [59] |
Zou B, Yang DL, Shi Z, et al. Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol, 2014, 165: 309-18. DOI:10.1104/pp.113.227801 |
| [60] |
Yang L, Wang Z, Hua J. Multiple chromatin‐associated modules regulate expression of an intracellular immune receptor gene in Arabidopsis. New Phytol, 2022, 237: 2284-97. |
| [61] |
Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Ann Rev Biochem, 2009, 78: 273-304. DOI:10.1146/annurev.biochem.77.062706.153223 |
| [62] |
Hopfner KP, Gerhold CB, Lakomek K, et al. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr Opin Struct Biol, 2012, 22: 225-33. DOI:10.1016/j.sbi.2012.02.007 |
| [63] |
Huang CY, Rangel DS, Qin X, et al. The chromatin-remodeling protein BAF60/SWP73A regulates the plant immune receptor NLRs. Cell Host Microbe, 2021, 29: 425-34. e4. DOI:10.1016/j.chom.2021.01.005 |
| [64] |
Zou B, Sun Q, Zhang W, et al. The Arabidopsis chromatin-remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy. Plant Cell Physiol, 2017, 58: 2202-16. DOI:10.1093/pcp/pcx155 |
| [65] |
Johnson KC, Xia S, Feng X, et al. The chromatin remodeler SPLAYED negatively regulates SNC1-mediated immunity. Plant Cell Physiol, 2015, 56: 1616-23. DOI:10.1093/pcp/pcv087 |
| [66] |
Halter T, Navarro L. Multilayer and interconnected post-transcriptional and co-transcriptional control of plant NLRs. Curr Opin Plant Biol, 2015, 26: 127-34. DOI:10.1016/j.pbi.2015.06.014 |
| [67] |
Lin J, Li QQ. Coupling epigenetics and RNA polyadenylation: missing links. Trends Plant Sci, 2023, 28: 223-34. DOI:10.1016/j.tplants.2022.08.023 |
| [68] |
Tsuchiya T, Eulgem T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc Natl Acad Sci U S A, 2013, 110: E3535-43. |
| [69] |
Marquez Y, Brown JW, Simpson C, et al. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res, 2012, 22: 1184-95. DOI:10.1101/gr.134106.111 |
| [70] |
Shen Y, Zhou Z, Wang Z, et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell, 2014, 26: 996-1008. DOI:10.1105/tpc.114.122739 |
| [71] |
Mandadi KK, Pyle JD, Scholthof KBG. Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C-3 and C-4 plant defenses. Mol Plant Microbe Interact, 2014, 27: 1277-90. DOI:10.1094/MPMI-05-14-0152-R |
| [72] |
Thatcher SR, Zhou W, Leonard A, et al. Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell, 2014, 26: 3472-87. DOI:10.1105/tpc.114.130773 |
| [73] |
Stamm S, Ben-Ari S, Rafalska I, et al. Function of alternative splicing. Gene, 2005, 344: 1-20. DOI:10.1016/j.gene.2004.10.022 |
| [74] |
Mandadi KK, Pyle JD, Scholthof KBG. Characterization of SCL33 splicing patterns during diverse virus infections in Brachypodium distachyon. Plant Signal Behav, 2015, 10: e1042641. DOI:10.1080/15592324.2015.1042641 |
| [75] |
Day IS, Golovkin M, Palusa SG, et al. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: insights into regulated splicing. Plant J, 2012, 71: 936-47. DOI:10.1111/j.1365-313X.2012.05042.x |
| [76] |
Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci U S A, 2000, 97: 1908-13. DOI:10.1073/pnas.020367497 |
| [77] |
Zhang XC, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol, 2007, 145: 1577-87. DOI:10.1104/pp.107.108720 |
| [78] |
Halterman DA, Wei FS, Wise RP. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol, 2003, 131: 558-67. DOI:10.1104/pp.014407 |
| [79] |
Costanzo S, Jia Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci, 2009, 177: 468-78. DOI:10.1016/j.plantsci.2009.07.012 |
| [80] |
Sela H, Spiridon LN, Petrescu AJ, et al. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol Plant Pathol, 2012, 13: 276-87. DOI:10.1111/j.1364-3703.2011.00744.x |
| [81] |
Cesari S, Thilliez G, Ribot C, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell, 2013, 25: 1463-81. DOI:10.1105/tpc.112.107201 |
| [82] |
Xu F, Xu S, Wiermer M, et al. The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes. Plant J, 2012, 70: 916-28. DOI:10.1111/j.1365-313X.2012.04906.x |
| [83] |
Sun B, Huang J, Kong L, et al. Alternative splicing of a potato disease resistance gene maintains homeostasis between growth and immunity. Plant Cell, 2024, 36: 3729-50. DOI:10.1093/plcell/koae189 |
| [84] |
Schweingruber C, Rufener SC, Zuend D, et al. Nonsense-mediated mRNA decay - Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta, 2013, 1829: 612-23. DOI:10.1016/j.bbagrm.2013.02.005 |
| [85] |
Rayson S, Arciga-Reyes L, Wootton L, et al. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One, 2012, 7: e31917. DOI:10.1371/journal.pone.0031917 |
| [86] |
Riehs-Kearnan N, Gloggnitzer J, Dekrout B, et al. Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res, 2012, 40: 5615-24. DOI:10.1093/nar/gks195 |
| [87] |
Gloggnitzer J, Akimcheva S, Srinivasan A, et al. Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe, 2014, 16: 376-90. DOI:10.1016/j.chom.2014.08.010 |
| [88] |
Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol, 2013, 64: 137-59. DOI:10.1146/annurev-arplant-050312-120043 |
| [89] |
Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell, 2007, 19: 2929-39. DOI:10.1105/tpc.107.051821 |
| [90] |
Zhai J, Jeong DH, De Paoli E, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev, 2011, 25: 2540-53. DOI:10.1101/gad.177527.111 |
| [91] |
Park JH, Shin C. The role of plant small RNAs in NB-LRR regulation. Brief Funct Genomics, 2015, 14: 268-74. DOI:10.1093/bfgp/elv006 |
| [92] |
Shivaprasad PV, Chen HM, Patel K, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell, 2012, 24: 859-74. DOI:10.1105/tpc.111.095380/elv006 |
| [93] |
Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood), 2003, 228: 111-33. DOI:10.1177/153537020322800201 |
| [94] |
Hubert DA, Tornero P, Belkhadir Y, et al. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J, 2003, 22: 5679-89. DOI:10.1093/emboj/cdg547 |
| [95] |
Lu R, Malcuit I, Moffett P, et al. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J, 2003, 22: 5690-9. DOI:10.1093/emboj/cdg546 |
| [96] |
Holt BF, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science, 2005, 309: 929-32. DOI:10.1126/science.1109977 |
| [97] |
Schulze-Lefert P. Plant immunity: the origami of receptor activation. Curr Biol, 2004, 14: R22-4. DOI:10.1016/j.cub.2003.12.017 |
| [98] |
Takahashi A, Casais C, Ichimura K, et al. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci U S A, 2003, 100: 11777-82. DOI:10.1073/pnas.2033934100 |
| [99] |
Bieri S, Mauch S, Shen QH, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 2004, 16: 3480-95. DOI:10.1105/tpc.104.026682 |
| [100] |
Muskett PR, Kahn K, Austin MJ, et al. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell, 2002, 14: 979-92. DOI:10.1105/tpc.001040 |
| [101] |
Azevedo C, Betsuyaku S, Peart J, et al. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J, 2006, 25: 2007-16. DOI:10.1038/sj.emboj.7601084 |
| [102] |
Boter M, Amigues B, Peart J, et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell, 2007, 19: 3791-804. DOI:10.1105/tpc.107.050427 |
| [103] |
Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell, 2006, 18: 491-501. DOI:10.1105/tpc.105.037234 |
| [104] |
Gou M, Shi Z, Zhu Y, et al. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J, 2012, 69: 411-20. DOI:10.1111/j.1365-313X.2011.04799.x |
| [105] |
Liu J, Huang Y, Kong L, et al. The malectin-like receptor-like kinase LETUM1 modulates NLR protein SUMM2 activation via MEKK2 scaffolding. Nat Plants, 2020, 6: 1106-15. DOI:10.1038/s41477-020-0748-6 |
| [106] |
Dong OX, Ao K, Xu F, et al. Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat Plants, 2018, 4: 699-710. DOI:10.1038/s41477-018-0216-8 |
| [107] |
Wang C, Zhu M, Hong H, et al. A viral effector blocks the turnover of a plant NLR receptor to trigger a robust immune response. EMBO J, 2024, 43: 3650-76. DOI:10.1038/s44318-024-00174-6 |
| [108] |
Zhang Y, Song G, Lal NK, et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun, 2019, 10: 3252. DOI:10.1038/s41467-019-11202-z |
| [109] |
Bigeard J, Rayapuram N, Pflieger D, et al. Phosphorylation- dependent regulation of plant chromatin and chromatin-associated proteins. Proteomics, 2014, 14: 2127-40. DOI:10.1002/pmic.201400073 |
| [110] |
Yamada K, Yamashita-Yamada M, Hirase T, et al. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J, 2016, 35: 46-61. DOI:10.15252/embj.201591807 |
| [111] |
Ma Y, Guo H, Hu L, et al. Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc Natl Acad Sci U S A, 2018, 115: 10218-27. DOI:10.1073/pnas.1811858115 |
| [112] |
Sarris PF, Duxbury Z, Huh SU, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell, 2015, 161: 1089-100. DOI:10.1016/j.cell.2015.04.024 |
| [113] |
Huh SU, Cevik V, Ding P, et al. Protein-protein interactions in the RPS4/RRS1 immune receptor complex. PLoS Pathog, 2017, 13: e1006376. DOI:10.1371/journal.ppat.1006376 |
| [114] |
Guo H, Ahn HK, Sklenar J, et al. Phosphorylation-regulated activation of the Arabidopsis RRS1-R/RPS4 immune receptor complex reveals two distinct effector recognition mechanisms. Cell Host Microbe, 2020, 27: 769-81.e6. DOI:10.1016/j.chom.2020.03.008 |
| [115] |
García AV, Parker JE. Heaven's gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci, 2009, 14: 479-87. DOI:10.1016/j.tplants.2009.07.004 |
| [116] |
Xu F, Cheng YT, Kapos P, et al. P-loop-dependent NLR SNC1 can oligomerize and activate immunity in the nucleus. Mol Plant, 2014, 7: 1801-4. DOI:10.1093/mp/ssu097 |
| [117] |
Mang HG, Qian W, Zhu Y, et al. Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell, 2012, 24: 1271-84. DOI:10.1105/tpc.112.096198 |
| [118] |
Zhu Y, Qian W, Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog, 2010, 6: e1000844. DOI:10.1371/journal.ppat.1000844 |
| [119] |
Jia M, Shen X, Tang Y, et al. A karyopherin constrains nuclear activity of the NLR protein SNC1 and is essential to prevent autoimmunity in Arabidopsis. Mol Plant, 2021, 14: 1733-44. DOI:10.1016/j.molp.2021.06.011 |
| [120] |
Palma K, Zhang Y, Li X. An importin α homolog, MOS6, plays an important role in plant innate immunity. Curr Biol, 2005, 15: 1129-35. DOI:10.1016/j.cub.2005.05.022 |
| [121] |
Chen H, Qian X, Chen X, et al. Cytoplasmic and nuclear Sw‐5b NLR act both independently and synergistically to confer full host defense against tospovirus infection. New Phytol, 2021, 231: 2262-81. DOI:10.1111/nph.17535 |
| [122] |
Wang Z, Liu X, Yu J, et al. Plasma membrane association and resistosome formation of plant helper immune receptors. Proc Natl Acad Sci U S A, 2023, 120: e2222-036120. |
| [123] |
Wu Z, Tian L, Liu X, et al. TIR signal promotes interactions between lipase-like proteins and ADR1-L1 receptor and ADR1-L1 oligomerization. Plant Physiol, 2021, 187: 681-6. DOI:10.1093/plphys/kiab305 |
| [124] |
Bi G, Su M, Li N, et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell, 2021, 184: 3528-41.e12. DOI:10.1016/j.cell.2021.05.003 |
| [125] |
Jacob P, Kim NH, Wu F, et al. Plant "helper" immune receptors are Ca2+-permeable nonselective cation channels. Science, 2021, 373: 420-5. DOI:10.1126/science.abg7917 |
| [126] |
Tang Y, Ho MI, Kang BH, et al. GBPL3 localizes to the nuclear pore complex and functionally connects the nuclear basket with the nucleoskeleton in plants. PLoS Biol, 2022, 20: e3001831. DOI:10.1371/journal.pbio.3001831 |
| [127] |
Li Y, Xue J, Wang FZ, et al. Plasma membrane-nucleo-cytoplasmic coordination of a receptor-like cytoplasmic kinase promotes EDS1-dependent plant immunity. Nat Plants, 2022, 8: 802-16. DOI:10.1038/s41477-022-01195-x |
| [128] |
Roth C, Lüdke D, Klenke M, et al. The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-α3 interaction partner required for plant immunity. Plant J, 2017, 92: 808-21. DOI:10.1111/tpj.13717 |
| [129] |
Liu X, Ao K, Yao J, et al. Engineering plant disease resistance against biotrophic pathogens. Curr Opin Plant Biol, 2021, 60: 101987. DOI:10.1016/j.pbi.2020.101987 |
| [130] |
Kourelis J, Marchal C, Posbeyikian A, et al. NLR immune receptor-nanobody fusions confer plant disease resistance. Science, 2023, 379: 934-9. DOI:10.1126/science.abn4116 |
| [131] |
Segretin ME, Pais M, Franceschetti M, et al. Single amino acid mutations in the potato immune receptor R3a expand response to phytophthora effectors. Mol Plant Microbe Interact, 2014, 27: 624-37. DOI:10.1094/MPMI-02-14-0040-R |
| [132] |
Huang H, Huang S, Li J, et al. Stepwise artificial evolution of an Sw-5b immune receptor extends its resistance spectrum against resistance-breaking isolates of Tomato spotted wilt virus. Plant Biotechnol J, 2021, 19: 2164-76. DOI:10.1111/pbi.13641 |
| [133] |
Xu G, Yuan M, Ai C, et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature, 2017, 545: 491-4. DOI:10.1038/nature22372 |
| [134] |
Kim JH, Castroverde CDM, Huang S, et al. Increasing the resilience of plant immunity to a warming climate. Nature, 2022, 607: 339-44. DOI:10.1038/s41586-022-04902-y |
 2025, Vol. 37
2025, Vol. 37 

 杨雷云,南京农业大学植物保护学院教授,国家高层次青年人才项目入选者,江苏省特聘教授。2020年,博士毕业于美国康奈尔大学;2020—2022年,在康奈尔大学从事博士后研究;2022年9月,加入南京农业大学植物保护学院稻麦真菌病害成灾机制与控制实验室,组建水稻稻瘟病抗性机制与遗传改良课题组。近年来,围绕植物免疫受体的鉴定和功能解析及免疫调控,以第一或通讯(含共同)作者在Plant Cell、New phytologist等期刊发表论文10余篇。担任Plant Cell期刊Assistant Features Editor,参与稿件审理和撰写亮点评述。主持或参与国家自然科学基金青年科学基金项目、国家自然科学基金重大项目、农业生物育种重大项目、国家重点研发项目、江苏特聘教授项目等;
杨雷云,南京农业大学植物保护学院教授,国家高层次青年人才项目入选者,江苏省特聘教授。2020年,博士毕业于美国康奈尔大学;2020—2022年,在康奈尔大学从事博士后研究;2022年9月,加入南京农业大学植物保护学院稻麦真菌病害成灾机制与控制实验室,组建水稻稻瘟病抗性机制与遗传改良课题组。近年来,围绕植物免疫受体的鉴定和功能解析及免疫调控,以第一或通讯(含共同)作者在Plant Cell、New phytologist等期刊发表论文10余篇。担任Plant Cell期刊Assistant Features Editor,参与稿件审理和撰写亮点评述。主持或参与国家自然科学基金青年科学基金项目、国家自然科学基金重大项目、农业生物育种重大项目、国家重点研发项目、江苏特聘教授项目等; 董莎萌,南京农业大学植物保护学院教授,教育部长江学者特聘教授,国家马铃薯产业技术体系岗位科学家。2008年,博士毕业于南京农业大学;2011—2014年,在英国The Sainsbury Laboratory从事博士后研究;2014年9月,加入南京农业大学植物保护学院作物疫病团队,组建晚疫病研究课题组。致力于推动农作物疫病研究从基础理论研究到前沿治理技术的创新:解释晚疫病菌的田间变异规律,揭示晚疫病的成灾机理;挖掘植物抗病资源;开展病害田间智能监控与绿色防控技术开发。在Science、PNAS学术权威杂志上发表SCI论文70余篇,论文总被引超过5 000次。主持国家自然科学基金重点项目、国家重点研发计划等国家级项目10余项,担任美国植物病理学会MPMI、英国植物病理学会MPP、中国植物学会JIPB、aBiotech等学术杂志编委
董莎萌,南京农业大学植物保护学院教授,教育部长江学者特聘教授,国家马铃薯产业技术体系岗位科学家。2008年,博士毕业于南京农业大学;2011—2014年,在英国The Sainsbury Laboratory从事博士后研究;2014年9月,加入南京农业大学植物保护学院作物疫病团队,组建晚疫病研究课题组。致力于推动农作物疫病研究从基础理论研究到前沿治理技术的创新:解释晚疫病菌的田间变异规律,揭示晚疫病的成灾机理;挖掘植物抗病资源;开展病害田间智能监控与绿色防控技术开发。在Science、PNAS学术权威杂志上发表SCI论文70余篇,论文总被引超过5 000次。主持国家自然科学基金重点项目、国家重点研发计划等国家级项目10余项,担任美国植物病理学会MPMI、英国植物病理学会MPP、中国植物学会JIPB、aBiotech等学术杂志编委







